Deliverable 11: D2.4: Transposon for converting any Gram-negative bacteria in an HFR strain
This work involves the description of a transposon able to mobilize an entire chromosome though conjugation between a donor and a DNA-less recipient. For this, all the mob/tra genes involved in conjugal transfer of super-promiscuous plasmid RP4 were retrieved from their original genetic frame and assembled as an artificial gene cluster in a mini-Tn5 transposon vector. Upon acquisition of such a transposon, any strain will acquire the ability of self-transference and therefore will behave as an HFR strain (High-frequency of recombination) that will ease the passing of the genomic information among strains with different degrees of new-to-nature reaction evolution.
Deliverable 2.1: Genome edited strains of E. coli and P. putida entirely cyborgized for hosting NTN reactions
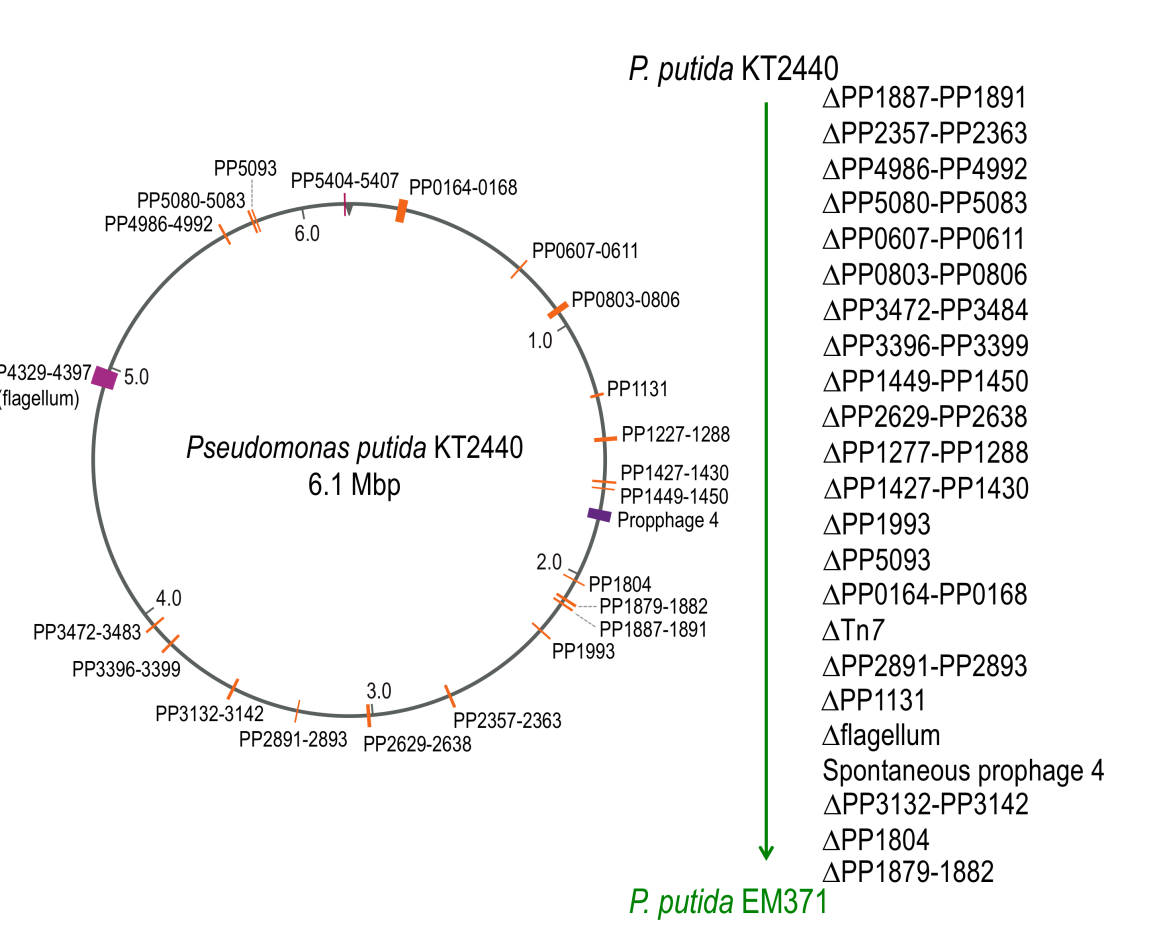
The natural reactions and metabolism of microorganisms can interfere with their use in research and industry. To optimize industrial processes, many initiatives try to erase as much genomic material as possible from these organisms, leaving only essential functions. In this deliverable, we describe our work on the reduction of the genomes of Escherichia coli and Pseudomonas putida, the two main bacterial species used as biological chassis. Our goal is to modify their cell membrane properties so that engineered products are externalized from the cell when they are produced, while maintaining the benefits of reduced genomes.
Deliverable 8 D3.2. Detailed report on genetically optimized artificial carboxylases based on evolved Sav isoforms
Metalloenzymes are important components of many metabolic pathways in living beings. Their metal-binding capabilities have made them the target of several scientific approaches. In the MADONNA Project, we are trying to combine these molecules with the biotin-streptavidin technology, a system that allows the isolation of the reaction in certain regions of the cell. Our goal is to develop a microbial cell factory that is able to fixate CO2 using artificial metalloenzymes, so that this gas can be used in the production of other chemicals. This process would reduce the emission of greenhouse gases while finding a sustainable alternative for their use.
Deliverable 13: D5.1 Morbidostat for heterotic computation of (bio)chemical problems
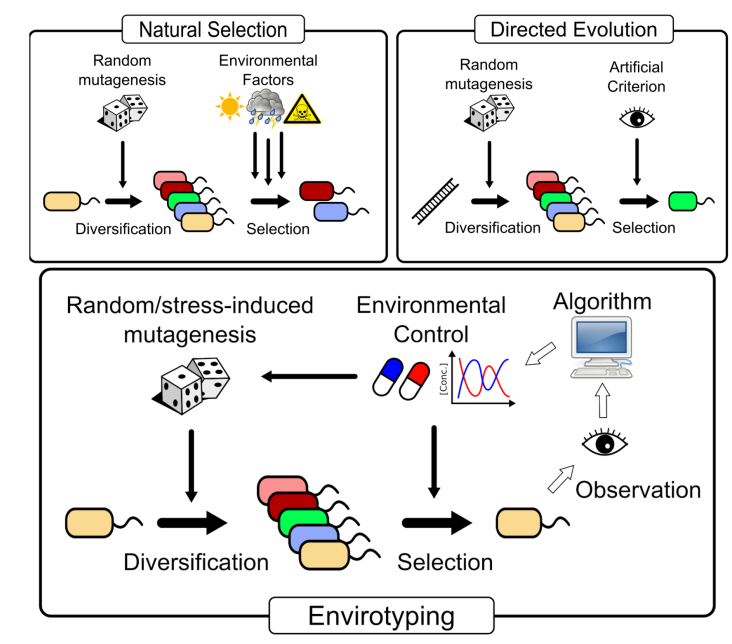
One way to boost evolutionary capabilities of organisms is to force them into stressful situations, thereby obliging them towards developing new solutions to survive. In the MADONNA project, we are studying this response by developing a special automated platform called a morbidostat. This platform monitors the growth of bacterial cultures and controls the addition of drugs so that the stressful stimuli is maintained. Then, the genomic material of these cultures will be analyzed to identify possible mutations. The morbidostat will be applicable to many experiments done as part of the MADONNA project.
Deliverable 14: D5.2 Automated generation of catalysts and information capture through a fitness function
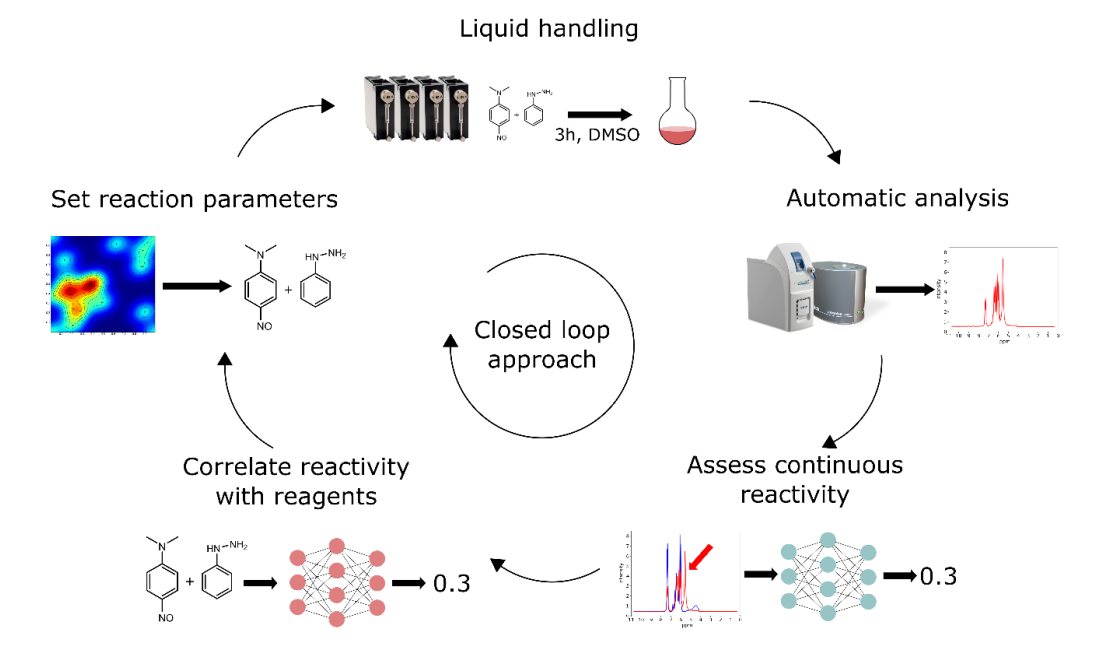
Automation and deep learning can become useful tools in the discovery of new chemical reactions, as they can speed up repetitive processes and analyse big datasets. Within the MADONNA project, we have developed a robotic platform that is able to identify reactions with high reactivity in a mixture, using nuclear magnetic resonance and mass spectrometry. Our goal was to create a close-loop framework where the device can determine the molecules involved in the reactions and set new reaction parameters in each analysis, performing an in-depth exploration of the chemical environment.
Deliverable 16: D5.4 Microdroplet-based environmental simulator
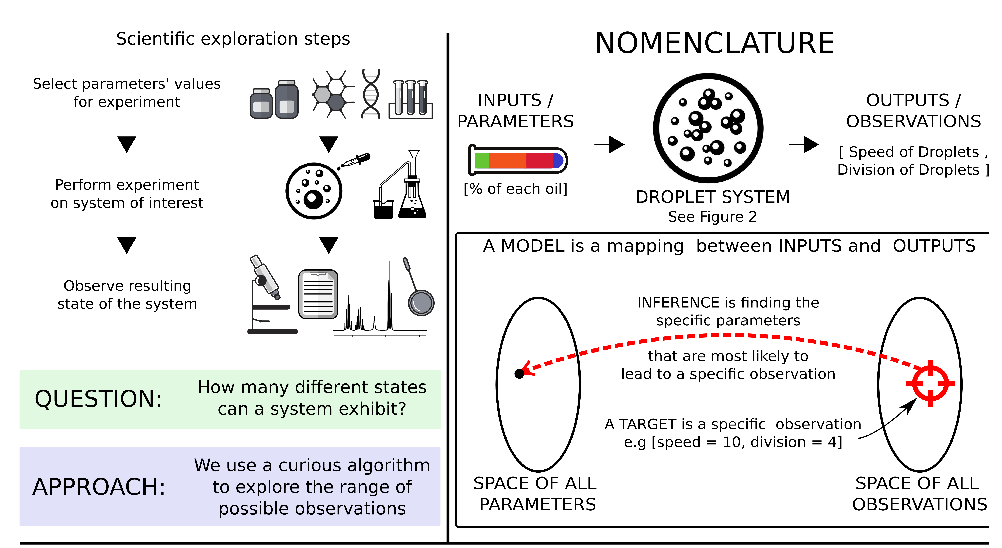
Robotic systems in laboratories are usually used in the optimization of known properties and depend on prior knowledge for their development. In this research, we have focused on the opposite approach: an autonomous system capable of extracting inputs from unknown environments. We have developed a chemical robot system which can identify specific parameters inside a sample that is most likely to lead to an observation. The device generates random inputs and changes external parameters so different states of the system can be studied along with their relation to their environment.
Deliverable 17: D6.1 An integrated computational frame for merging NTN reactions to extant metabolism
The use of microorganisms for the removal of contaminants is a promising approach taken by many biotechnological industries. Here, we have developed mathematical and computational models aimed at understanding and improving bioremediation systems. Our main use cases include: 1) the coupling of plastic waste generation and a community of virtual microorganisms to study differences found in sample studies of ocean microplastic abundance and 2) a model used to explore microbial bioremediation networks inside their metabolism with the goal of boosting their capabilities.
Deliverable 18: D6.2 Proposition of containment needs for microbial agents bearing new reactions
A common topic in biotechnological discussions is the risk of harmful outcomes for the local ecosystems due to the presence of modified microorganisms despite the lack of scientific evidence to support most of these conclusions. Commonly, containment is achieved by “genetic firewall” – the process of modifying microbes so that they cannot interact with other life forms around them. However, some interesting functions including many bioremediation approaches, can only be accomplished when all environment is involved. Within the MADONNA project, we have studied alternative firewall methods and tested them virtually using computational models to better understand the relation between engineered microbes and their propagation capabilities.